Toolbar
Toolbar icons are shortcuts to the most common PySisyphe functions.
Icons displayed in toolbar can be modified in the settings dialog box (tab General > Toolbar icons, left-click on the  icon to toggle visbility).
icon to toggle visbility).

Display a file selection dialog box to open a PySisyphe volume (.xvol). You can also use menubar (File > Open…), or double-clicking on an empty space in the thumbnail bar, or select the thumbnail bar popup menu “open volume”, or press the keyboard shortcut CTRL-O (CMD-O MacOS platform).

Save the selected volume of the thumbnail bar (blue border), displayed in a view widget. You can also use menubar (File > Save…), or select the thumbnail popup menu “Save”, or press the keyboard shortcut CTRL-S (CMD-S MacOs platform).

Save all open volumes in the thumbnail bar. You can also use menubar (File > Save all), or select the thumbnail bar popup menu Save all volume(s).

Close the selected volume of the thumbnail bar (blue border). You can also use menubar (File > Close), or press DEL key.

Close all open volumes in the thumbnail bar. You can also use menubar (File > Close all), or the thumbnail bar popup menu Close all volume(s).

Display a dialog box to edit attributes (identity, acquisition, image) of the selected volume in the thumbnail bar (blue border). You can also use menubar (File > Edit attributes…), or press the keyboard shortcut CTRL-? (CMD-? MacOs platform).

Display a dialog box to copy a reference volume ID to a list of volume(s), ROI(s), meshs(es) or streamlines. You can also use menubar (File > ID replacement…).

Display a dialog box to anonymize a list of volume(s). You can also use menubar (File > Anonymize…), or select the thumbnail popup menu Anonymize.

Display a dialog box to edit labels of the the selected volume in the thumbnail bar (blue border). This function is assigned to a label volume with LB modality. In this volume, each scalar value in the image array is associated with a label. These labels are written to a xml file with the same name as the volume and .xlabels extension. You can also use menubar (File > Edit volume labels…).

Display a dialog box to import Minc volume(s) (.mnc). You can also use menubar (File > Import > Import Minc…).

Display a dialog box to import Nifti volume(s) (.img, .img.gz, .hdr, .nii, .nii.gz, .nia). You can also use menubar (File > Import > Import Nifti…).

Display a dialog box to import Nrrd volume(s) (.nrrd). You can also use menubar (File > Import > Import Nrrd…).

Display a dialog box to import old format Sisyphe volume(s) (.vol). You can also use menubar (File > Import > Import Sisyphe…).

Display a dialog box to import VTK volume(s) (.vtk, vti). You can also use menubar (File > Import > Import VTK…).

Display a file selection dialog box to open a FreeSurfer volume (.mgh). You can also use menubar (File > Open from format… > Open FreeSurfer MGH…).

Display a file selection dialog box to open a Minc volume (.mnc). You can also use menubar (File > Open from format… > Open Minc…).

Display a file selection dialog box to open a Nifti volume (.img, .img.gz, .hdr, .nii, .nii.gz, .nia). You can also use menubar (File > Open from format… > Open Nifti…).

Display a file selection dialog box to open a Nrrd volume (.nrrd). You can also use menubar (File > Open from format… > Open Nrrd…).

Display a file selection dialog box to open a BrainVoyager volume (.vmr). You can also use menubar (File > Open from format… > Open BrainVoyager VMR…).

Display a file selection dialog box to open an old format Sisyphe volume (.vol). You can also use menubar (File > Open from format… > Open Sisyphe…).

Display a file selection dialog box to open an VTK volume (.vtk, .vti). You can also use menubar (File > Open from format… > Open VTK…).

Save the selected volume in the thumbnail bar (blue border) in Nifti format (.nii). You can also use menubar (File > Save to format… > Save Nifti…).

Save the selected volume in the thumbnail bar (blue border) in Numpy format (.npy). You can also use menubar (File > Save to format… > Save Numpy…).

Save the selected volume in the thumbnail bar (blue border) in Nrrd format (.nrrd). You can also use menubar (File > Save to format… > Save Nrrd…).

Save the selected volume in the thumbnail bar (blue border) in Minc format (.mnc). You can also use menubar (File > Save to format… > Save Minc…).

Save the selected volume in the thumbnail bar (blue border) in VTK format (.vtk). You can also use menubar (File > Save to format… > Save VTK…).

Display a dialog box to import Dicom files. You can also use menubar (File > DICOM > DICOM Import…).

Display the selected volume of the thumbnail bar (blue border) in the slice view. If no thumbnail is selected, the first volume to the left of the thumbnail bar is selected and displayed in the slice view. You can also select thumbail popup menu Display in slice view, or press F1 key.

Display the selected volume of the thumbnail bar (blue border) in the orthogonal view. If no thumbnail is selected, the first volume to the left of the thumbnail bar is selected and displayed in the orthogonal view. You can also select thumbail popup menu Display in orthogonal view, or press F2 key.

Display the selected volume of the thumbnail bar (blue border) in the synchronized view. If no thumbnail is selected, the first volume to the left of the thumbnail bar is selected and displayed in the synchronized view. You can also select thumbail popup menu Display in synchronized view, or press F3 key.
Display the selected volume of the thumbnail bar (blue border) in the projection view. If no thumbnail is selected, the first volume to the left of the thumbnail bar is selected and displayed in the projection view. You can also select thumbail popup menu Display in projection view, or press F4 key.

Display the selected volume of the thumbnail bar (blue border) in the multi-component view. If no thumbnail is selected, the first volume to the left of the thumbnail bar is selected and displayed in the multi-component view. This viewer is assigned to multi-component volume (array image values are vectors, not scalar values). You can also select thumbail popup menu Display in multi-component view, or press F5 key.
Join several single-component volumes to produce a multi-component volume. Display a file selection dialog box to open a list of single-component volumes. You can also use menubar (Functions > Join single component volume(s)…).

Break down a multi-component volume into single-component volumes. Display a file selection dialog box to open multi-component volumes. You can also use menubar (Functions > Split multi component volume(s)…).
Display a dialog box to flip volume axes (x-axis left/right, y-axis anterior/posterior, z-axis cranial/caudal). You can also use menubar (Functions > Flip axis…).
Display a dialog box to permute volume axes. This function is used to correct axes order i.e. coronal to axial conversion or sagittal to axial conversion. You can also use menubar (Functions > Permute axis…).

Display a dialog box to edit attributes (modality, sequence, scalar value unit, frame, set default origin and set default directions) of a volume list. You can also use menubar (Functions > Attributes conversion…).

Display a dialog box for building or editing lookup tables. You can also use menubar (File > Edit LUT…).

Display a dialog box to perform texture features. You can also use menubar (Functions > Texture feature maps…).

Display a dialog box to process mean filtering. You can also use menubar (Functions > Filters > Mean…).

Display a dialog box to perform median filtering. You can also use menubar (Functions > Filters > Median…).

Display a dialog box to perform gaussian filtering. You can also use menubar (Functions > Filters > Gaussian…).

Display a dialog box to perform gradient magnitude. You can also use menubar (Functions > Filters > Gradient magnitude…).

Display a dialog box to perform gradient laplacian. You can also use menubar (Functions > Filters > Laplacian…).

Display a dialog box to perform anisotropic diffusion filtering. You can also use menubar (Functions > Filters > Anisotropic diffusion…).

Display a dialog box to perform bias field filtering. You can also use menubar (Functions > Bias field correction…).

Display a dialog box for voxel by voxel. You can also use menubar (Functions > Voxel-by-voxel processing > Algebra…).

Display a dialog box to automate a series of processings. You can also use menubar (Functions > Workflow processing…).

Display a dialog box for detecting fiducial markers of a Leksell stereotaxic frame. You can also use menubar (Registration > Stereotacitc frame detection…).
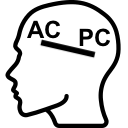
Display a dialog box to select anterior and posterior commissure of a volume. You can also use menubar (Registration > AC-PC selection…).

Display a dialog box to reorient a volume and/or change the field-of-view. You can also use menubar (Registration > Volume reorientation…).

Display a dialog box to perform a manual co-registration of two volumes. You can also use menubar (Registration > Manual registration…).
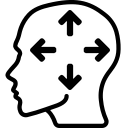
Display a dialog box to perform a rigid co-registration of two volumes. You can also use menubar (Registration > Rigid registration…).
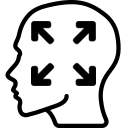
Display a dialog box to perform an affine co-registration of two volumes. You can also use menubar (Registration > Affine registration…).
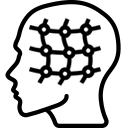
Display a dialog box to perform an non linear displacement field-based co-registration of two volumes. You can also use menubar (Registration > Displacement field registration…).

Display a dialog box to perform an ICBM152 spatial normalization of a volume. You can also use menubar (Registration > ICBM normalization…).
Display a dialog box to perform an asymmetry analysis of a volume. You can also use menubar (Registration > Asymmetry displacement field…).
Display a dialog box for processing the Jacobian determinant of a displacement field. Assigned to the multi-component volume and displacement field sequence attribute. You can also use menubar (Registration > Displacemlent field jacobian determinant…).

Display a dialog box to perform a skull stripping of a volume. This function make use of pre-trained deep learning models. You can also use menubar (Segmentation -> skull stripping…).

Display a dialog box to perform prior-based tissue segmentation of a volume (three to six classes: cortical gray matter, subcortical gray matter, white matter, cerebro-spinal fluid, brainstem, cerebellum). You can also use menubar (Registration > Tissue segmentation…).

Toggle between full-screen and normal display modes. In full-screen mode, the central area of the current visualization widget (sectional view, orthogonal view, synchronized view, projection view, multi-component view) is extended to the full screen. Press F11 or ESC key to exit full-screen mode.

Display a dialog box for editing PySisyphe preferences. You can also use menubar (File > preferences…).

Close PySisyphe. You can also use menubar (File > Quit…).